R2DT documentation
What is R2DT?
The R2DT software automatically generates RNA secondary structure diagrams in consistent, reproducible and recognisable layouts using a library of templates representing a wide range of RNAs.
Try it now
How does it work?
Find out more about R2DT, read the R2DT paper, or check out the slides from a recent talk:
Examples
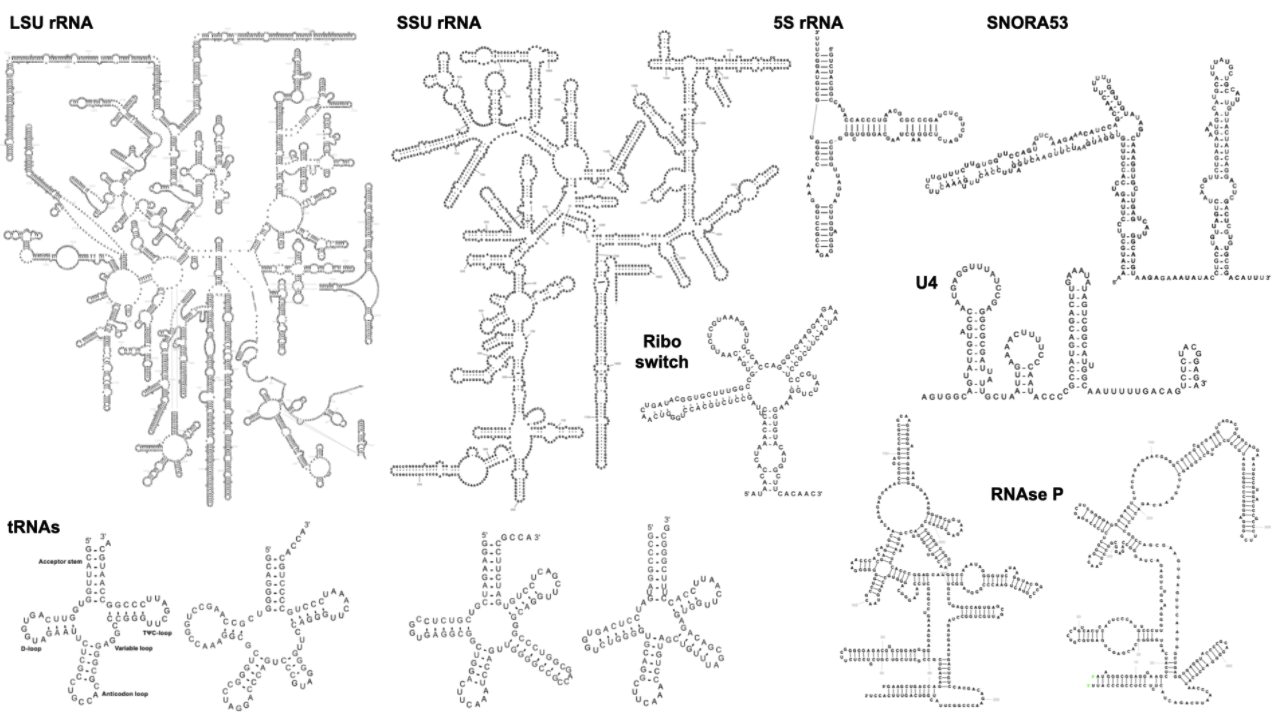
The following example visualisations show LSU, SSU, and 5S rRNA, four tRNAs, two RNAse P, snoRNA, MoCo riboswitch, and U4 snRNA:
Getting started
R2DT can be used in a number of ways:
Web application hosted by RNAcentral
API powered by EMBL-EBI Web Services (see API and Python package)
As a command line tool with Docker, Podman, Singularity, or as bare metal installation
Who uses R2DT?
RNAcentral uses R2DT to visualise >20 million RNA secondary structures
PDBe uses R2DT to enable interactive navigation between sequence, 2D and 3D structure (for example, 1S72)
Rfam and GtRNAdb display R2DT diagrams in sequence similarity search
NAKB uses R2DT to visualise secondary structure of RNAs from PDB (for example, 1S72).
Browse papers citing R2DT →
Citation
If you use R2DT in your work, please consider citing the following paper:
R2DT is a framework for predicting and visualising RNA secondary structure using templates Nature Communications
License
R2DT is available under the Apache 2.0 license.
Get in touch
If you have any questions or feedback, feel free to submit a GitHub issue or contact the RNAcentral help desk.
GitHub
Contribute to the R2DT project on GitHub →

